
Proteros Gallery Assays (based on Reporter Displacement Assay)
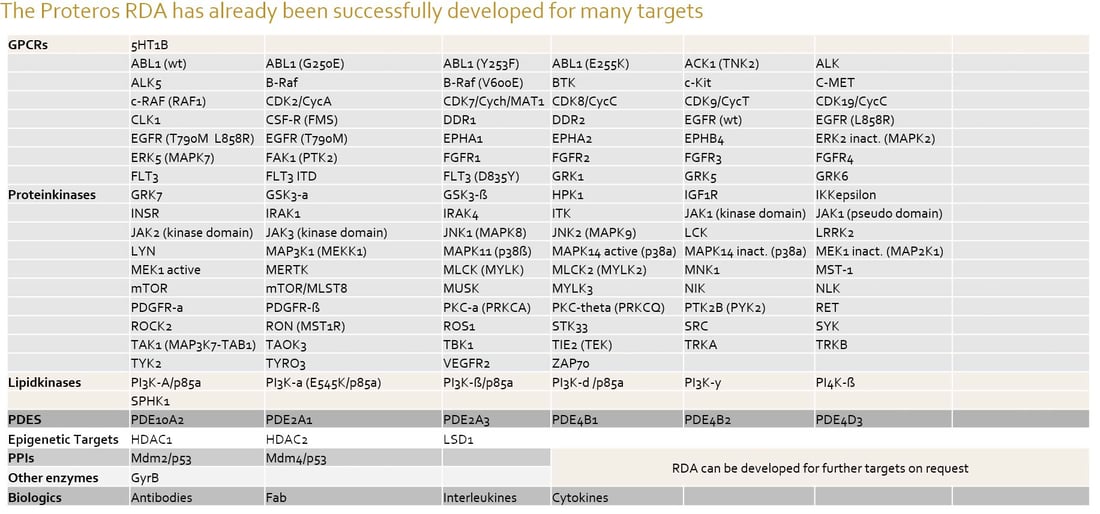
Gallery Assays - Proteros has established Reporter Displacement Assays for > 100 protein targets.
The RDA Gallery assays are applicable for library screening, affinity profiling and kinetic profiling immediately.
- The RDA can be used to profile ligands as fragments, small molecules, peptides, protein-protein-interactions and biologics
- Reporter probes are designed from small molecules, peptides, DNA, co-factors, protein binding partners (PPI) and biologics. The RDA can be used for soluble proteins and membrane proteins.
- The RDA represents a truly high throughput binding assay
- Reporter probes are designed that bind to the target site of interest
- Reporter probe binding to the target leads to a specific optical signal that can be read in 384 well format
- When the ligand of interest binds to the target, the reporter probe is displaced, which leads to a dose dependent loss of the reporter probe binding signal
- At equilibrium, compound affinity can be determined (Kd). Monitoring the time dependent signal loss yields the compound’s binding kinetics (koff, kon and the residence time for non-covalent ligands and kinact for covalent ligands)
Gallery Assays - The Proteros reporter displacement assay (RDA)
Why use Proteros Assays, Biophysics and Screening
Assay development and screening
Expertise to match assay and biophysics strategy to required insights
Screening and profiling technologies
Full suite of biophysical and biochemical screening and profiling analysis platforms
Access to fit-for-purpose screening libraries
High-quality small molecule and fragment screening libraries
Agility and flexibility of execution
Dedicated, expert project management and proactive communication